Download
Abstract
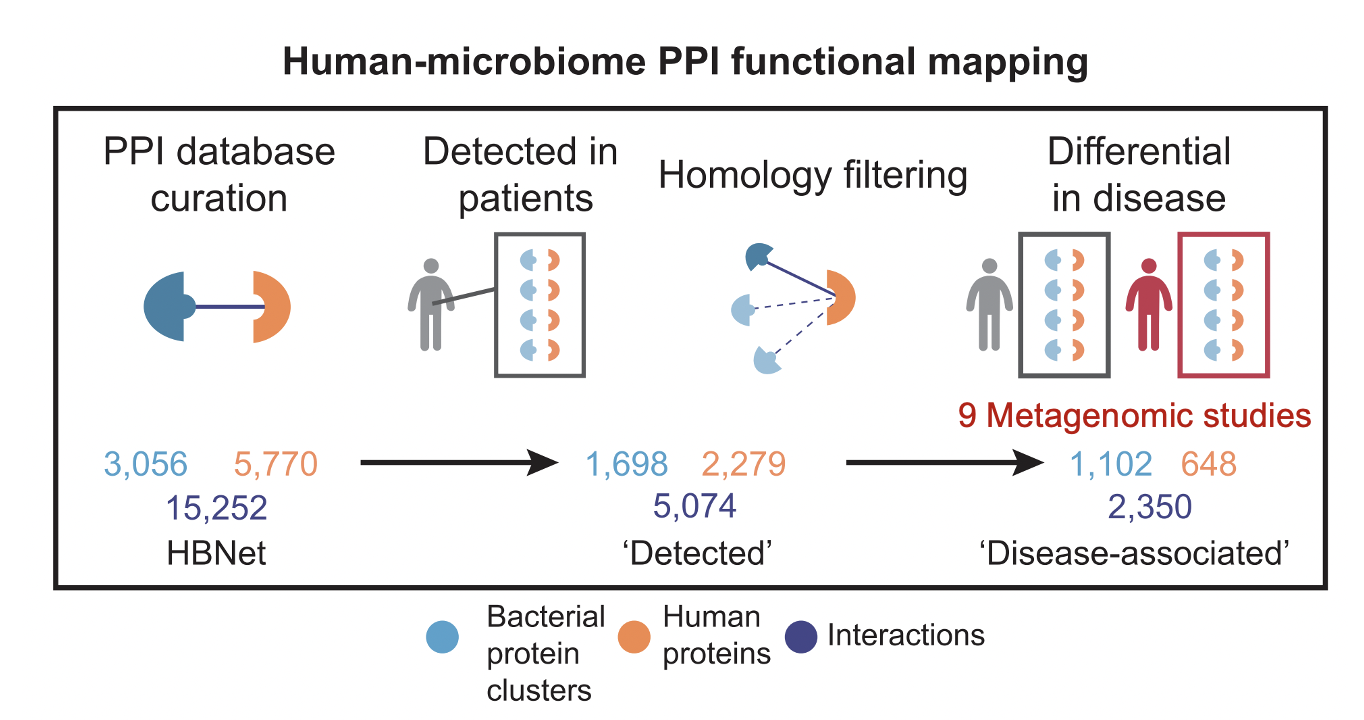
To identify potential pathways through which human-associated bacteria impact host health, we leverage publicly-available interspecies protein-protein interaction (PPI) data to find clusters of microbiome-derived proteins with high sequence identity to known human-protein interactors. We observe differential targeting of putative human-interacting bacterial genes in nine independent metagenomic studies, finding evidence that the microbiome broadly targets human proteins involved in immune, oncogenic, apoptotic, and endocrine signaling pathways in relation to IBD, CRC, obesity, and T2D diagnoses. This host-centric analysis provides a mechanistic hypothesis-generating platform and extensively adds human functional annotation to commensal bacterial proteins.
Citation
Zhou, Hao, Juan Felipe Beltrán, and Ilana Lauren Brito. “Host-microbiome protein-protein interactions capture disease-relevant pathways.” Genome Biology 23.1 (2022): 72.
@article{zhou2022host,
title={Host-microbiome protein-protein interactions capture disease-relevant pathways},
author={Zhou, Hao and Beltr{\'a}n, Juan Felipe and Brito, Ilana Lauren},
journal={Genome Biology},
volume={23},
number={1},
pages={72},
year={2022},
publisher={Springer}
}